From the Journal of Clinical Microbiology, Leinberger et al uses microarray technology to rapidly speciate human fungal pathogens.
Susceptibility testing is already quite unreliable for bacteria, and it is even less useful for fungal infections. In fact antifungal agent choice is mostly based on the speciation of the fungus. Yet speciation is typically performed by examining the shape of the fungus and performing biochemical tests on fungal colonies. This requires a relatively large number of fungi, which will needs a couple of days to grow. This process is of course the rate-limiting step of clinical decision making in treating fungal infections.
Using DNA microarray technology, Leinberger et al produced a spot array chip containing unique sequences of rRNAs from 12 common pathogenic fungi. These were validated against 21 speciated clinical isolates. They found that the microarray chip was able to rapidly (within about 4 hours) and accurately speciate 12 of the most common fungal pathogens in the genus Candida and Aspergillus.
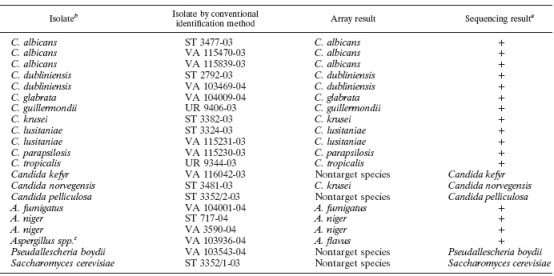
This clinical applications of DNA microarray technology is particularly clinically relevant, as critically ill and immunocompromised patients can be infected by multiple fungal species. Rapid and targeted antifungal selection should help clinicians better balance the treatment of infections against the often serious adverse effects of antifungals.
Technorati Tags:
Medicine / Statistics / Computational biology / Fungal speciation / Gene expression microarray




No comments:
Post a Comment